
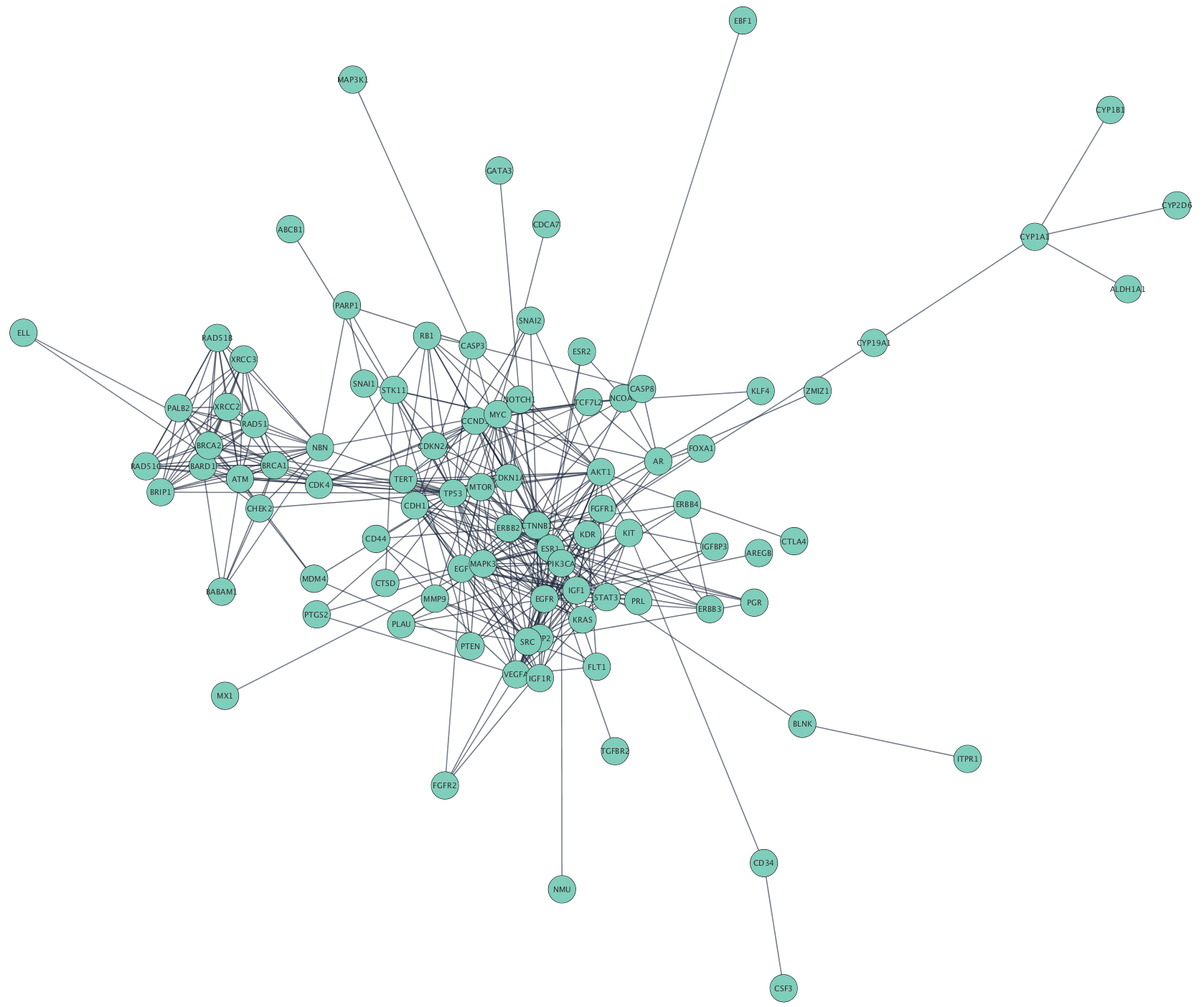
For example, in figure 1a nodes 2, 3, 4 form a clique of size 3. The course will also access and analyse the data through Cytoscape apps, including the new IntAct app. Network Analysis with Cytoscape Tutorial, November 2013.
#Cytoscape 3 tutorial software
The course will focus on giving attendees hands-on experience in the use of Cytoscape, an open source software platform for complex network analysis and visualization. We will subsequently use these networks to overlay large-scale data such as that obtained through RNA-Seq or mass-spec proteomics.
#Cytoscape 3 tutorial how to
Attendees will learn how to construct protein-protein interaction networks and explore them in full detail. This course provides an introduction to the basic theory and concepts of network analysis. We aim to simulate the classroom experience as closely as possible, with opportunities for one-to-one discussion with tutors and a focus on interactivity throughout. PLEASE NOTE The Bioinformatics Team are presently teaching as many courses live online.
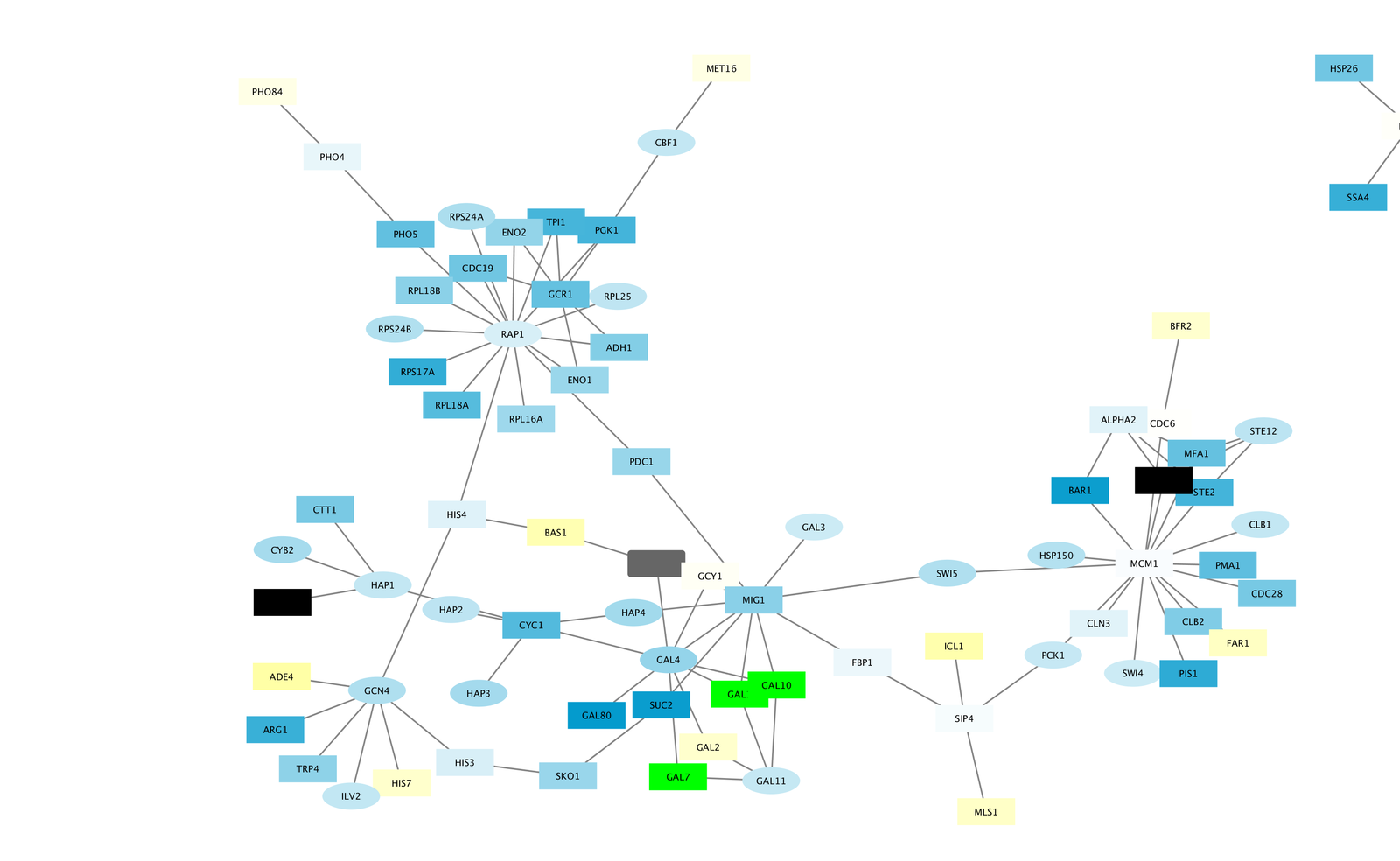
Office of Student Conduct, Complaints and Appeals.Judge Business School Entrepreneurship Centre.Institute of Continuing Education Staff Learning & Development.Engineering Centre for Languages & Inter-Communication.Development and Alumni Relations - Staff Learning & Development.Department of Physics - Health & Safety.Cambridge University Library Staff Learning & Development.Cambridge Centre for Teaching and Learning.gWidgets_0.0-52 RGtk2_2.20.25 RCurl_1.95-4.- Select training provider - ( Bioinformatics ) stats graphics grDevices utils datasets methods Tutorials for Cytoscape 2.x Tutorial 1: Prepare RIN data Tutorial 2: Use node sets Tutorial 3: Analyze (un)weighted networks Tutorial 4: Compare RINs. Note- you don’t necessarily need the GUI and can instead get the working functions from the following repos: You should see a window that looks like this: At the top of the Cytoscape 3. Cytoscape 3 User Interface Launch Cytoscape 3. This tutorial describes the Cytoscape 3 user interface. Comparing my and your session it looks like the error is caused by your lack of: gWidgetsRGtk2_0.0-81. Handout IntroductiontoCytoscape3.pdf 2 Cytoscape is an open source software tool for integrating, visualizing, and analyzing data in the context of networks. the target column is 2 and the weight column is 3.
#Cytoscape 3 tutorial install
I just upgraded to R 3.0.0 and, running the code in question “source(“”)” on a fresh install gives me no problems. We will explain how to display this network with NeAT, Cytoscape, yED and VisANT.


 0 kommentar(er)
0 kommentar(er)
